Dr Maria Górna from the CNBCh UW, Faculty of Chemistry and her team developed a new method of preparing RNA for sequencing, which will facilitate the study of gene expression for many species of fungi, plants and eukaryotic microorganisms.
The Structural Biology Group, led by Dr Górna, studies the properties of human antiviral IFIT proteins. The team has developed new methods of selecting and enriching RNA, covered by a patent application filed in Poland, the USA and the EU.
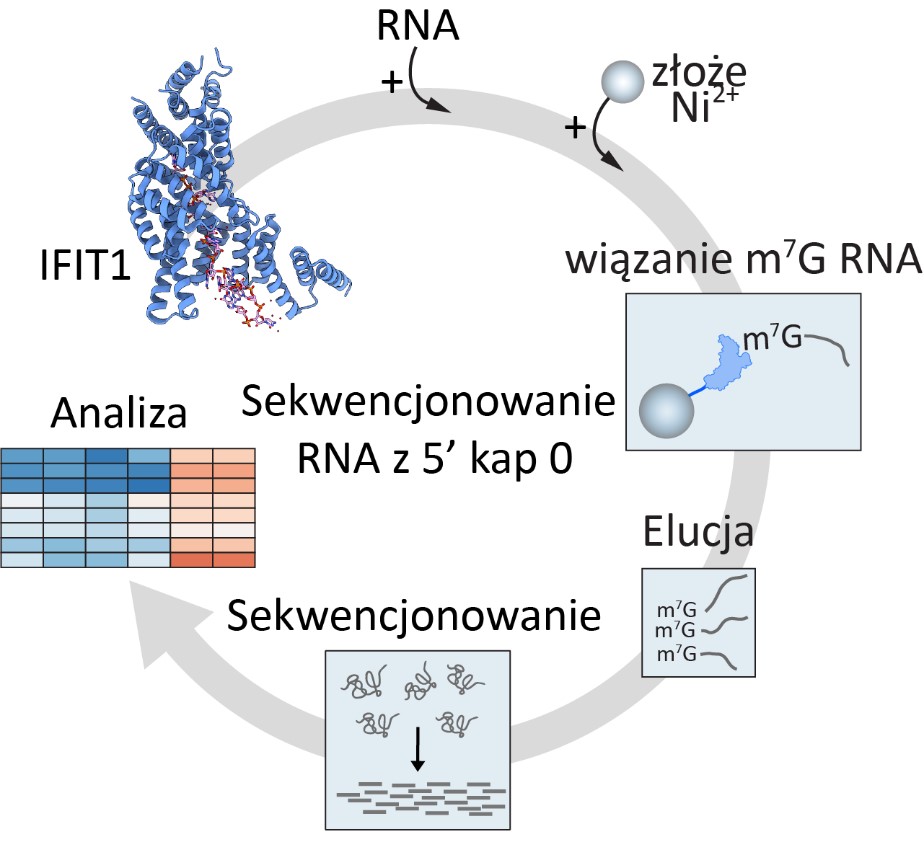
The publication in the prestigious “Nucleic Acids Research” journal describes the first application of this invention to improve the sequencing of coding RNA using the example of baker’s yeast – a model organism in the study of gene expression and RNA metabolism. Dr Górna’s method consists in capturing and enriching the coding RNA by binding (of the IFIT1 protein immobilized on the resin) to the cap group at the 5 ‘end of the RNA – the so-called cap 0, which is present only at the ends of the mRNA. Non-coding RNAs are washed away and removed from the sample, thereby improving the quality of the sequencing data.
The analysis of the RNA enriched in this way was carried out in collaboration with the Laboratory of Translatomics IBB PAS under the supervision of Prof. Agata Starosta and proved that the new method successfully replaces commercial kits used for the preparation of mRNA samples. In addition, Dr Agnieszka Tudek from the Institute of Biochemistry and Biophysics, the Polish Academy of Sciences, and Prof. Rafał Tomecki from the Faculty of Biology, the University of Warsaw, showed that the previously known method based on another cap-binding protein is not suitable for yeast RNA samples.
This work in Dr Górna’s group was supported – among others – by the LIDER grant from the National Centre for Research and Development, Poland, and the EMBO Installation Grant.
Publication details
Martyna Nowacka, Przemysław Latoch, Matylda A. Izert, Natalia K. Karolak, Rafał Tomecki, Michał Koper, Agnieszka Tudek, Agata L. Starosta, Maria W. Górna, A cap 0-dependent mRNA capture method to analyze the yeast transcriptome, “Nucleic Acids Research”, 2022, DOI: 10.1093/nar/gkac903.
Source: www.uw.edu.pl