Laboratory of Epitranscriptomics
About the Group:
Viruses represent one of the greatest threats to human health and the global economy. While their impact on human health is relatively well understood, our knowledge of host-pathogen interactions in other mammals and animals remains limited. Evolutionary differences and the diverse ecological niches of various species make it difficult to directly extrapolate findings from human cells to other animals. Since livestock serve as a key source of protein for humans, understanding how these animals defend themselves against pathogens is critical for maintaining global food security.
Moreover, viruses that cause infectious diseases in livestock can often jump to new hosts, including humans, potentially leading to epidemics or even pandemics.
Research in the Laboratory of Epitranscriptomics focuses on understanding how innate immunity combats infections caused by RNA viruses, such as influenza virus, SARS-CoV-2, Dengue virus, and Ebola virus. Specifically, we investigate how host cell receptors recognize viral RNA and how viruses evade detection by masking their RNA to escape the cellular defense system
Research activities:
Scientific projects:
• Deciphering how viral epitranscriptome shapes host immune response
National Science Centre, Sonata Bis (2022-2026), PI Paweł Sikorski
• Novel versatile tool for RNA labeling
Excellence Initiative – Research University New Ideas (IDUB Nowe Idee) (2023-2025), PI Paweł Sikorski
• FLU-SWITCH Identification of factors driving the emergence and spread of avian influenza viruses with zoonotic potential
Project coordinated by Dr. Romain Volmer, INRAE; polish partner: Paweł Sikorski.
ERA-NET ICRAD (International Coordination of Research on Infectious Animal Diseases) (2023-2026)
Publications:
- Drazkowska K., Cieslicka J., Kitowicz M., Pastucha A., Markiewicz L., Szymanek W., Goryca K., Kowalczyk T., Cysewski D., Bausch A.R., Sikorski P.J. (2025) Effective recognition of double-stranded RNA does not require activation of cellular inflammation. Science Advances, 11(15).
- Gackowska K., Krejmer-Rabalska M., Drazkowska K., Jemielity J., Krol E., Szewczyk B. (2025) Affinity-purified sHBsAg-based virus-like particles as a platform for foreign mRNA binding. Virology, 110651.
- Drazkowska K., Tomecki R., Tudek A. (2025) Purification of Enzymatically Active Xrn1 for Removal of Non-capped mRNAs from In Vitro Transcription Reactions and Evaluation of mRNA Decapping Status In Vivo. Methods in Molecular Biology. 2863: 81-105.
- Tomecki R., Drazkowska K., Madaj R., Mamot A., Dunin-Horkawicz S., Sikorski P.J. (2024) Expanding the Available RNA Labeling Toolbox With CutA Nucleotidyltransferase for Efficient Transcript Labeling with Purine and Pyrimidine Nucleotide Analogs. ChemBioChem. 25(15): e202400202
- Warminski M., Trepkowska E., Smietanski M., Sikorski P.J., Baranowski M.R., Bednarczyk M., Kedzierska H., Majewski B., Mamot A., Papiernik D., Popielec A., Serwa R.A., Shimanski B.A., Sklepkiewicz P., Sklucka M., Sokolowska O., Spiewla T., Toczydlowska-Socha D., Warminska Z., Wolosewicz K., Zuberek J., Mugridge J.S., Nowis D., Golab J., Jemielity J., Kowalska J. (2024) Trinucleotide mRNA Cap Analogue N6-Benzylated at the Site of Posttranscriptional m6Am Mark Facilitates mRNA Purification and Confers Superior Translational Properties In Vitro and In Vivo. Journal of the American Chemical Society. 146(12): 8149-8163.
- Tomecki R., Drazkowska K., Kobylecki K., Tudek A. 2023. SKI complex: A multifaceted cytoplasmic RNA exosome cofactor in mRNA metabolism with links to disease, developmental processes, and antiviral responses. Wiley Interdisciplinary Reviews RNA. e1795.
- Drazkowska K., Tomecki R., Warminski M., Baran N., Cysewski D., Depaix A., Kasprzyk R., Kowalska J., Jemielity J., Sikorski P.J. 2022. 2′-O-Methylation of the second transcribed nucleotide within the mRNA 5′ cap impacts the protein production level in a cell-specific manner and contributes to RNA immune evasion. Nucleic Acids Research. 50: 9051–9071.
Research Equipment:
| Equipment Name | Contact Person / Room Number | |
| 1 | Reversed Phase-High Performance Liquid Chromatography (RP-HPLC) | Pawel Sikorski, pawelsikorski@uw.edu.pl, room: 2.104 |
| 2 | BSL2 laminar flow cabinets | Pawel Sikorski, pawelsikorski@uw.edu.pl, room: 2.104 |
| 3 | CO2 incubators | Pawel Sikorski, pawelsikorski@uw.edu.pl, room: 2.104 |
Team Leader:
Paweł Sikorski, PhD, DSc, completed his doctoral studies at the Faculty of Biology, University of Warsaw (supervised by Prof. Joanna Kufel). He then pursued postdoctoral training in Prof. Jacek Jemielity’s group at the Center for New Technologies, University of Warsaw. Since 2022, he has been an assistant professor at the Faculty of Biology, University of Warsaw, additionally he leads the Laboratory of Epitranscriptomics at Biological and Chemical Research Centre, University of Warsaw.
In 2023, he was awarded his habilitation degree and received both the Individual Prize of the Rector of the University of Warsaw and the Prime Minister of Poland Award for his scientific achievements encompassing his habilitation dissertation.
He also completed an international research internship in the laboratory of Dr. Dominique Gagliardi at the IBMP in Strasbourg, France. Moreover, Dr. Sikorski is the author of over 20 scientific publications in prestigious journals such as Nucleic Acids Research, Journal of the American Chemical Society, and Angewandte Chemie, as well as four international patent applications.
Significant Achievements:
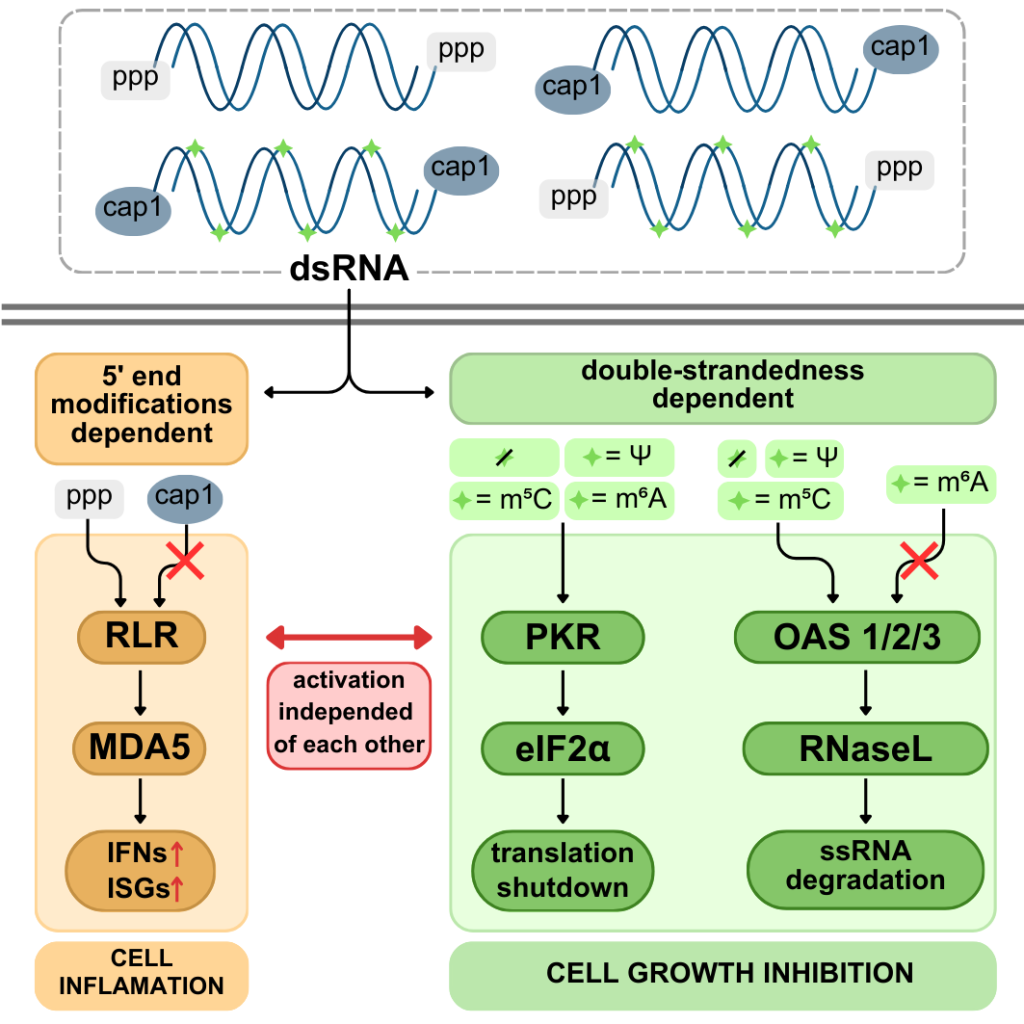
Excess double-stranded RNA (dsRNA) in human cells, often following viral infections, activates innate immune pathways that trigger inflammation and inhibit cell growth. We showed that dsRNA responses can occur without inflammation. The pro-inflammatory RLR pathway and the growth-inhibitory OAS/RNase L- and PKR pathways function independently, with 5’ dsRNA ends driving inflammation and RNA duplex structures activating OAS/RNase L and PKR. Unexpectedly, common RNA modifications – N6-methyladenosine (m6A), 5-methylcytosine, and pseudouridine – did not affect dsRNA immunogenicity overall. However, m6A specifically inhibited the OAS/RNase L pathway. Our findings reveal how innate immunity is finely tuned to counter specific threats.